Arbalet
Table of Contents
- What is Arbalet ?
- Download
- Turorial
- File formats
- How can we compare trees
- More information
- Bug report
What is Arbalet ?
Arbalet is a Java tool to view and manipulate phylogenetic trees and compute their Parsimony score.
With Arbalet, given a file of taxa and a tree in Newick format you can:
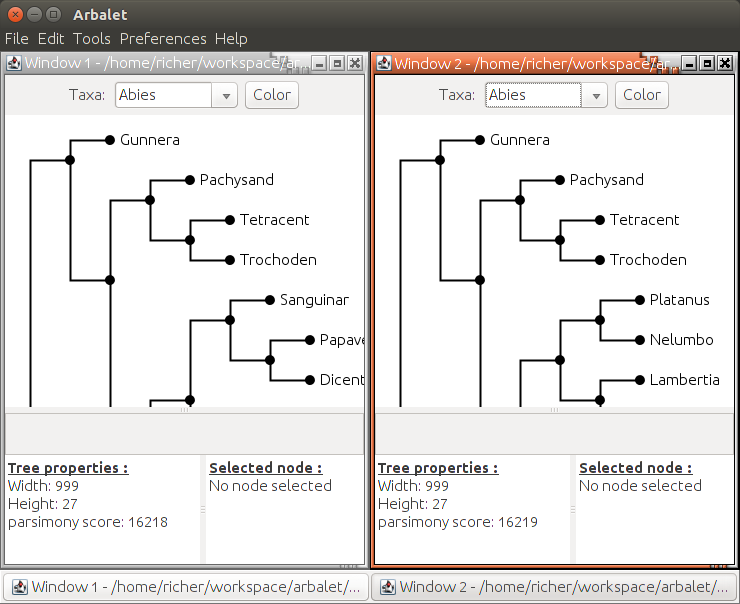
- visualize a parsimony tree and the score of its nodes
- Scroll and zoom to examine some part of the trees
- Export the tree as an image (PNG, SVG, ...)
- Compare two trees in terms of number of operations to transform one tree to another
Download Arbalet
You can download the following version:
- arbalet-1.0.jar (compiled with jdk 1.7.0_55)
Examples with one file of sequences (zilla) and two trees in newick format:
- taxa file: zilla.sg (500 taxa)
- newick tree 16218: zilla_16218.newick
- newick tree 16219: zilla_16219.newick
Source code is available on sourceforge.net
Tutorial
Please follow this link toget access to the tutorial.
File formats
For sequences you can use one of the following format:
- Phylip (.phy)
5 10 S1 AACCTTTTATTA S2 CA-CTTTTATTA S3 GGGTTTTTATTA S4 TGG?TTTTATTA S5 TACCTGTTATTA - Fasta (.fasta)
>S1 AACCTT TTATTA >S2 CA-CTT TTATTA >S3 GGGTTT TTATTA >S4 TGG?TT TTATTA >S5 TACCTG TTATTA - SequenceGroup (.sg), name of sequence followed by residues
S1 AACCTTTTATTA S2 CA-CTTTTATTA S3 GGGTTTTTATTA S4 TGG?TTTTATTA S5 TACCTGTTATTA
For trees, use the Newick format:
How can we compare trees ?
Ribeiro and Vianna implementation
We use a derivative form of Path-Relinking (PR) introduced by Glover and Laguna (see CiteSeerX). PR is a Metaheuristic (see Wikipedia for a definition) with an intensification strategy used to explore elite solutions.
PR consider two solutions called source and guiding solutions and consists in transforming the source solution into the guiding solution by applying a series of modifications. After each modification the current solution is duplicated and an exploration phase is performed on a copy of the solution.
If we remove the exploration phase we then have a modification algorithm from one tree to another.
For the Maximum Parsimony problem an implementation was given by Ribeiro and Vianna. This implementation can be called top-down implementation as it starts from the root of the tree and recursively explore each left and right subtrees. For each iteration we compare the taxa in the left and right subtrees of the source and guiding solution. All taxa of the source left (resp. right) subtree that are in the right (resp. left) subtree of the guiding solution are moved to the right (resp. left) subtree of the of the source solution.
The major drawback of this implementation is that it requires a lot of modifications and moves of the taxa from left to right (resp. right to left) subtrees. When a taxon is degraphed it is then regraphed on a branch of the sibling subtree that minimizes the parsmony score of the tree. We can also put the degraphed taxon at the top of the sibling subtree to avoid the search of a minimum tree.
Richer and Vazquez-Ortiz implementation
We have implemented a bottom-up implementation which compares the subtrees present in the source and guiding solutions and starts with subtrees of size 1, size 2, then size 3 and so on. When a subtree (X,Y) of the guiding solution is not found in the source solution then we modify the source tree by degraphing Y and regraphing it on X.
For example, with two trees of zilla (500 taxa) of score 16218 and 16219 we have:
- TD1: top-down (implementation of Ribeiro and Vianna) with regraph on root of sibling subtree: 462 transformations
- TD2: top-down with minimization on regraph (implementation of Ribeiro and Vianna) with regraph on branch that minimizes the overall score of the tree: 343 transformations
- BU: bottom-up (implementation of Richer and Vazquez-Ortiz): 32 transformations.
Here are some more results:
| guiding / source | BU Transform. |
BU Time (s) |
TD1 Transform. |
TD1 Time (s) |
TD2 Transform. |
TD2 Time (s) |
| 16218 / 162182 | 24 | 0.10 | 118 | 0.33 | 74 | 0.85 |
| 16218 / 16219 | 32 | 0.14 | 462 | 1.36 | 343 | 4.08 |
| 16218 / 16250 | 97 | 0.40 | 1779 | 4.79 | 1519 | 25.32 |
| 16218 / 16401 | 151 | 0.60 | 1891 | 5.16 | 1474 | 25.36 |
| 16218 / 21727 | 446 | 1.68 | 1881 | 5.17 | 1241 | 18.70 |
As we can see the Bottom-Up (BU) implementation requires a smaller number of transformations to transform one tree into another. The execution time is the most important for TD2 because we need to find the score of each regraph on every branch and keep the one that minimizes the parsimony score of the tree.
More information
Arbalet is an acronym that comme from Arbre (the french term for tree) and alet (the contraction of applet) as it was initially supposed to be an Applet.
The contributors to this project are:
- BEUTIER Kévin, student of the Master 2 SILI (Solutions Informatique LIbres)
- BRUNEAU-VOISINE Florian, student of the Master 2 SILI
- GUILLERMIN Clément, student of the Master 2 SILI
- LERAY Marc, student of the Master 2 SILI
- VAZQUEZ-ORTIZ Karla Esmeralda, PhD Student
- RICHER Jean-Michel, Associate Professor, LERIA, University of Angers
Kévin, Florian, Clément and Marc did the main job, we have added some minor (but interesting) capabilities to the final project (like tree comparison with Path-Relinking, search and visualisation of a taxon in the tree) and killed some bugs.